About
This server performs cosegregation analysis by the Full-Likelihood method [1] and outputs a Bayes factor that can be integrated into the variant classification guidelines developed by the American College of Medical Genetics and Genomics (ACMG) and the Association of Molecular Pathology (AMP).
The website is designed for classifying variants in a known disease gene as pathogenic or benign within the context of clinical genetic testing. If you want to conduct cosegregation analysis for gene discovery research, i.e., to find out which gene associates with the disease of interest, then please see my other package VICTOR.
Quick start
Click on the “ANALYSIS” tab, provide a gene symbol and upload a pedigree file, then click the “Submit” button.
Pedigree File
The Pedigree File must be a tab- or space-delimited text file. If you edit data in Excel, please save as “Tab delimited Text (.txt)”. Missing values are represented by a period. Empty fields, space within fields, or quoted fields are not allowed. IDs are case-sensitive with alphanumeric characters or the _ character.
Required columns:
PedID: Pedigree_ID. Alphanumeric. It cannot be 0. Multiple pedigrees are allowed.
IndID: Individual_ID. Alphanumeric. It cannot be 0. IndID does not need to be unique across pedigrees.
Father: Individual_ID of father. Put 0 for founders. If one parent is 0, both parents must be 0.
Mother: Individual_ID of mother. Put 0 for founders. If one parent is 0, both parents must be 0.
Sex: Biological sex, not the personal identification of one's own gender. 0=UnknSex, 1=M=Male, 2=F=Female.
Aff*: Affection status. Value is UnknAff (unknown), Unaff (unaffected), or a disease name.
Age: Age-of-onset for affected and age-of-the-last-exam for unaffected individuals. 0 for unknown age.
FPTP: The first person tested positive for the variant in each pedigree. 1 for FPTP; 0 for others.
Geno: Genotype. 0=unknown, neg=negative, het=heterozygous-carrier, hom=homozygous-carrier. You don’t need to input genotype for obligatory carriers as the program will infer automatically. You don’t need to input negative genotype (non-carrier) for spouses even if you assume that the variant enters the pedigree only once. The program will fill in the genotype based on allele frequency.
Optional columns:
Twin: Twin status. 0: not twin; positive integer: siblings with the same number are twins.
Pop: Population. This column overrides the advanced option "Population" on the webpage.
Cohort: Year range. This column overrides the advanced option "Year range" on the webpage.
Comment: Information to be shown in pedigree drawings.
* For cancer-associated genes, Aff is the first diagnosis of any cancer. Below is a list of strings for the “Aff”:
------------------------------------------------------------------------------------------------------------------------
Name Descriptions
------------------------------------------------------------------------------------------------------------------------
unaff unaffected
Lip Lip
Tongue Tongue
Mouth Mouth
Oral Oral cavity (lip, tongue, mouth)
Saliv Salivary gland
Parotid Parotid gland
Tonsil Tonsil
Oroph Oropharynx
Nasoph Nasopharynx
Pyrifm Pyriform sinus
Hypoph Hypopharynx
Pharynx Pharynx (include Oropharynx, Nasopharynx, Hypopharynx, Pharynx unspecified)
BCP Buccal cavity and pharynx (include Lip, Tongue, Mouth, Saliv, Parotid, Tonsil, Pharynx)
Throat Oropharynx, Tonsil, Base of tongue
Nasal Nasal cavity and middle ear
A.sinus Accessory sinuses
Larynx Larynx
Trachea Trachea
Oesoph Oesophagus
Stomach Stomach (synonym: Gastric)
SmBowel Small intestine
Colon Colon
RS.junc Rectosigmoid junction
Rectum Rectum
CRC Colorectal cancer (include Colon, RS.junc, Rectum)
Anus Anus
Liver Liver
Gall Gallbladder
Biliary biliary tract
PanCa Pancreas
BilPan biliary tract and Pancreas
Lung Lung
Thymus Thymus
Heart Heart
Bone Bone
Bone.l Bone of limbs
Bone.o Bone other than limbs
Osteo Osteosacoma
Sarcoma Soft tissue sarcoma and bone sarcoma
CM Cutaneous Melanoma
NM.skin Non-melanoma of skin
Meso Mesothelioma
STS Soft-tissue sarcoma (Mesothelioma, Kaposi, Peripheral nerves, Peritoneum & retroperitoneum, Connective & soft tissue)
BrCa Breast
Vagina Vagina
Cervix Cervix uteri
Corpus Corpus uteri
Uterus Cervix or corpus uteri
OvCa Ovary
Penis Penis
ProCa Prostate
Testis Testis
UpUrin Upper Urinary tract malignancy (kidney and renal pelvis)
Kidney Kidney
RenalCC Renal Cell Carcinoma (only for CI5-XI, a subset of UpUrin)
RCC Renal Cell Carcinoma (only for CI5-XI, a subset of UpUrin)
Ureter Ureter
Bladder Bladder
Urinary Urinary tract (include Kidney, Renal pelvis, Ureter, Bladder, Other urinary organs)
Eye Eye
UM Uveal melanoma
Mening Meninges
CNS Central nervous system
Brain Brain
Thyroid Thyroid
MTC Medullary Thyroid Cancer
Adrenal Adrenal gland
Hodgkin Hodgkin lymphoma
NH.lym Non-Hodgkin lymphoma
IPD Immunoproliferative diseases
Myeloma Multiple myeloma
L.leuk Lymphoid leukemia
M.leuk Myeloid leukemia
U.leuk Cell-unspecified leukemia
Leuk Leukemia
Lymph Lymphoid Neoplasms (include Hodgkin lymphoma, Non-Hodgkin lymphoma, MALT-lyphoma, Lymphoid leukaemia)
------------------------------------------------------------------------------------------------------------------------
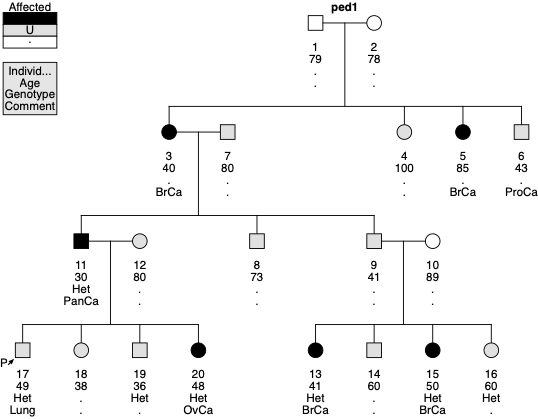
Below is an example Pedigree File and a drawing for BRCA1. Please note that affection status for analysis depends on the gene. Because the liability class model for BRCA1 involves only breast, ovarian, and pancreatic cancer, individual 6 (prostate cancer) and 17 (lung cancer) are “unaffected”. Therefore, the squares for these two persons are not filled with solid black color. Also note that the proband, individual 17, is not affected. This is correct as the definition of proband is the first person who tested positive for the variant.
PedID IndID Father Mother Sex Aff Age Geno FPTP
ped1 1 0 0 M . 79 . 0
ped1 2 0 0 F . 78 . 0
ped1 3 1 2 F BrCa 40 . 0
ped1 4 1 2 F unaff 100 . 0
ped1 5 1 2 F BrCa 85 . 0
ped1 6 1 2 M ProCa 43 . 0
ped1 7 0 0 M unaff 80 . 0
ped1 8 7 3 M unaff 73 . 0
ped1 9 7 3 M unaff 41 . 0
ped1 10 0 0 F . 89 . 0
ped1 11 7 3 M PanCa 30 Het 0
ped1 12 0 0 F unaff 80 . 0
ped1 13 9 10 F BrCa 41 Het 0
ped1 14 9 10 M unaff 60 . 0
ped1 15 9 10 F BrCa 50 Het 0
ped1 16 9 10 F unaff 60 Het 0
ped1 17 11 12 M Lung 49 Het 1
ped1 18 11 12 F unaff 38 . 0
ped1 19 11 12 M unaff 36 Het 0
ped1 20 11 12 F OvCa 48 Het 0
-
1.Thompson D, Easton DF, Goldgar DE. A Full-Likelihood Method for the Evaluation of Causality of Sequence Variants from Family Data. The American Journal of Human Genetics. 2003;73(3):652-655.
-
2.Kuchenbaecker KB et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA. 2017;317(23):2402-2416. PMID:28632866.
-
3.Antoniou AC et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer. 2008;98(8):1457-66. PMID:18349832.
-
4.Mocci E et al. Risk of pancreatic cancer in breast cancer families from the breast cancer family registry. Cancer Epidemiol Biomarkers Prev. 2013;22(5):803-11. doi: 10.1158/1055-9965.EPI-12-0195. PMID:23456555.
-
5.BOADICEA V7 Release 114.
-
6.Belman et al. Considerations in assessing germline variant pathogenicity using cosegregation analysis. Genetic in Medicine. 2020