Contents
Classification of germline variants within disease-causing genes as pathogenic or benign is crucial for the translation of clinical genetic testing results. Cosegregation analysis of pedigree data (also called causality analysis) is a useful tool for assessing germline variant pathogenicity. This server performs cosegregation analysis by the Full-Likelihood method [1] and outputs a Bayes factor that can be integrated into a multifactorial variant classification scheme. It can also be transformed into a strength category to be used in the application of the variant classification guidelines developed by the American College of Medical Genetics and Genomics (ACMG) and the Association of Molecular Pathology (AMP). This server provides penetrance for 16 cancer-associated genes. It can also analyze other cancer genes if you provide a relative risk table, or other non-cancer genes if you provide penetrance. The website makes pedigree drawings from an input pedigree file.
The website is designed for classifying variant in a known disease gene as pathogenic or benign within the context of clinical genetic testing. If you want to conduct cosegregation analysis for a gene discovery research, i.e., to find out which gene associates with the disease of interest, then please see my other package VICTOR.
Now I have put COOL in two servers, Amazon Web Services and University of Utah Center for High-Performance Computing. They should be identical. If one server is down, please use the other one. Later when the website is stable, I will shutdown the Amazon server.
The simplest way to do a cosegregation analysis is to provide a gene symbol and a pedigree file. The gene symbol must be one of the genes for which this server provides penetrance. For a quick tutorial, (1) copy the below Pedigree File example and save to a file in your computer; (2) click "Choose File" to upload the Pedigree File; (3) type BRCA1 in the "Gene Symbol" box; (4) click "Submit".
The cosegregation analysis results will be written to another web page; the URL of which will be shown on the screen. The result page contains parameters, Bayes factors and pedigree drawings. Results will be stored on the server for 30 days. After each analysis, you can click “show the previous page” (Safari) or “go back” (Chrome, FireFox) to do another analysis.
Long text will be truncated in the pedigree drawing. However, if you hover your mouse over the individual symbol, the comment will be shown on the screen. You can save the result page as a webarchive or print it to a PDF. It is recommended to save as webarchive because it will keep the mouse-hovering feature. The webarchive contains pedigree drawings as .svg files. You can open and modify those files by Adobe Illustrator.
Allele frequency: Allele frequency of the variant of interest from databases such as gnomAD. Otherwise, program will use the default 0.0001.
Break loops: If the program complains about loops, check your Pedigree File. If the loops are real, input yes and then run analysis again. The program will break loops for you. Loop can be due to consanguinity (e.g., someone married a cousin) or marriage (e.g., two brothers married two sisters).
Population: One or more population names divided by a comma. Alternatively, it can be NFE, LAT, AFR, SAS, EAS, or FIN, which stands for non-Finish European, Latino, African, South Asian, East Asian, or Finish, respectively. This option is not required if you use the Comprehensive Pedigree Format (see below).
Year range: It can be 1993-1997 or 1998-2002 or 2003-2007 or 2008-2012. This option is not required if you use the Comprehensive Pedigree Format (see below).
Variant of interest: If your Pedigree File is in Comprehensive Pedigree Format (see below), it may contain genotypes of multiple variants. This option allows you to choose which variant to analyze.
The Pedigree File must be a text file. If you edit data in Excel, please save as “Tab delimited Text (.txt)”. The recommended file format is called Comprehensive Pedigree Format (CPF). This format is designed to contain all necessary information for cosegregation analysis, while eliminating ambiguity (e.g. discrepancy in affection status assignment or proband definition) and improving usability (i.e., directly analyzable by COOL) and re-usability (i.e., re-analyze an old file without modifications when we change the disease risk model or the gene of interest). I hope it helps to streamline the analysis process and facility data sharing. If CPF is too difficult, you can use the Freestyle Format below. The server can read both formats.
In addition to performing cosegregation analysis, you can use this website to check for problems in a CPF file. If a computation is not successful, error message will be shown on the screen. Even if computation is successful, you can scroll down to the bottom of the result page to look for warnings about the file.
If your gene of interest is a cancer-associated gene and the penetrance is not provided by this website, you can upload a Relative Risk File. The website will calculate penetrance from data in your Relative Risk File and the Cancer Incidence in 5 Continents (CI5).
This file contains relative risk for multiple genes. The file can be divided into sections; one section for one gene. Between sections, blank lines or comment lines (starting with #) will be ignored. Each gene section starts with a "--gene=<GennSymbol>" line and ends with a "TableEnd" line. Between them is a relative risk table; one row for one age group and one column for a genotype, sex, and cancer site combination. The first three rows are headers for genotype, sex, and disease, respectively. The first column defines age group. Each gene can associate with an unlimited number of diseases. Please see “Supported Cancer Sites” below for the allowed disease names. List the most prevalent disease on the left-most column. Age group is defined by two numbers: the first can be 0/5/10/15/20/25/30/35/40/45/50/55/60/65/70/75/80/85; the second can be 4/9/14/19/24/29/34/39/44/49/54/59/64/69/74/79/84/+.
For dominant genes, add the --dominant line right after --gene=<GennSymbol>, then you don't need the "hom" columns. For recessive genes, you only need the "hom" columns, skipping the "het" columns. The --crhf option specifies the cumulative risk haplotype carrier frequency in the population. When this option is set, the program will calculate the non-carrier and carrier incidence rates so that the combined incidence, based on this carrier frequency, is equal to the observed population incidence in CI5.
Analysis by this website can use a model in which each family member is followed-up until the diagnosis of first cancer, risk-reducing treatment, or the last observation, whichever occurred first. This can be specified using the options “--ce-file=” or “--censoring+=”. The former will read a file for earliest censoring events such as cancers or risk-reducing treatments. The website provides two files, coseg2.ce.cancer.txt and coseg2.ce.HBOC.txt. The first one includes all cancer sites. The second one includes all cancer sites plus prophylactic mastectomy and prophylactic oophorectomy that are relevant to BRCA1/2. If you want to define your own censoring events, then use the “--censoring+=” option. Please notice the “+” sign. It means you can use this option many times, each time adding new events to the list. Arguments of this option can be one event or multiple events separated by a comma (,).
The Relative Risk File must be a text file. If you edit data in Excel, please save as “Tab delimited Text (.txt)”.
Below is an example Relative Risk File:
# BRCA1/2 Female BrCa & OvCa from Kuchenbaecker KB et al. JAMA. 2017;317(23):2402-2416. [2]
# BRCA1/2 Male BrCa from Antoniou AC et al. Br J Cancer. 2008;98(8):1457-66. [3]
# BRCA1/2 PanCa from Mocci E et al. Cancer Epidemiol Biomarkers Prev. 2013;22(5):803-11. [4]
# BRCA2 ProCa from BOADICEA (V7 Release 114) [5]
--gene=BRCA1
--crhf=0.00075
--ce-file=coseg2.ce.HBOC.txt
Geno het het het het het het hom hom hom hom hom hom
Sex Male Male Male Female Female Female Male Male Male Female Female Female
Disease BrCa OvCa PanCa BrCa OvCa PanCa BrCa OvCa PanCa BrCa OvCa PanCa
0-29 8 1 4.68 73.7 1 4.68 8 1 4.68 73.7 1 4.68
30-39 8 1 4.68 46.2 41.4 4.68 8 1 4.68 46.2 41.4 4.68
40-49 8 1 4.68 17.2 56.7 4.68 8 1 4.68 17.2 56.7 4.68
50-59 8 1 1.40 9.7 53.3 1.40 8 1 1.40 9.7 53.3 1.40
60-69 8 1 1.40 7.0 69.1 1.40 8 1 1.40 7.0 69.1 1.40
70-79 8 1 1.40 4.8 11.8 1.40 8 1 1.40 4.8 11.8 1.40
TableEnd
--gene=BRCA2
--dominant
--crhf=0.0013
--ce-file=coseg2.ce.HBOC.txt
Geno het het het het het het het het
Sex Male Male Male Male Female Female Female Female
Disease BrCa OvCa PanCa ProCa BrCa OvCa PanCa ProCa
0-29 80 1 4.77 7.33 60.8 1 4.77 1
30-39 80 1 4.77 7.33 20.3 7.3 4.77 1
40-44 80 1 4.77 7.33 16.4 15.9 4.77 1
45-49 80 1 4.77 7.33 16.4 15.9 4.77 1
50-54 80 1 2.03 7.33 11.4 24.5 2.03 1
55-59 80 1 2.03 7.33 11.4 24.5 2.03 1
60-64 80 1 2.03 7.33 6.4 21.5 2.03 1
65-69 80 1 2.03 3.39 6.4 21.5 2.03 1
70-74 80 1 2.03 3.39 6.6 4.4 2.03 1
75-79 80 1 2.03 3.39 6.6 4.4 2.03 1
80-84 1 1 1 3.39 1 1 1 1
TableEnd
Penetrance File is useful for non-cancer genes, or when you want to use your own incidence data instead of those from CI5. But you have to calculate a liability class for each individual and make sure the liability class in the Pedigree File matches with the lines in the Penetrance File.
If you have N liability classes, the first line in the Penetrance File is the number N followed by some options about the liability class model including --genes, --diseases, --age-div, --unkn-aoo, and --unkn-sex. The programs will check whether the number N matches the option --age-div and --diseases. Suppose your --age-div argument has 6 numbers, they divide ages into 6+1=7 age groups. Suppose the --gene is associated with 2 --diseases, then you have (2+1)*7=21 liability classes for one sex, and N=21*2=42 liability classes in total.
After the first line are N lines of penetrance. Each line has 3 numbers corresponding to the penetrance for non-carriers, heterozygous carriers, and homozygous carriers, respectively. After the numbers are description of the liability class. For unaffected class, the description should contain the phrase “unaff”. Line order is important too. The first few lines are penetrance for unaffected individuals, then affected individuals in the same order as the --diseases argument. Within each unaffected and affected section, lines are sorted by sex (male to female) and age group (young to old).
Below is an example Penetrance File for BRCA2 in standard format. This file is currently in use by this web tool by default. Please see Reference for the source of penetrance for female breast and ovarian cancer [2], male breast cancer [3], pancreatic cancer [4], and prostate cancer [5].
The Penetrance File must be a text file. If you edit data in Excel, please save as “Tab delimited Text (.txt)”.
120 << --age-div=30,40,45,50,55,60,65,70,75,80,85 --gene=BRCA2 --diseases=BrCa,OvCa,PanCa,ProCa --rr=coseg_rr.txt --bing=Yes
0.000007279 0.000092860 0.000092860 << 1 unaff male 0-29 UK 1998-2002
0.000056313 0.000660008 0.000660008 << 2 unaff male 30-39 UK 1998-2002
0.000183361 0.002015465 0.002015465 << 3 unaff male 40-44 UK 1998-2002
0.000558795 0.005528094 0.005528094 << 4 unaff male 45-49 UK 1998-2002
0.001815873 0.015685099 0.015685099 << 5 unaff male 50-54 UK 1998-2002
0.005487972 0.042679786 0.042679786 << 6 unaff male 55-59 UK 1998-2002
0.014030174 0.101554466 0.101554466 << 7 unaff male 60-64 UK 1998-2002
0.030272736 0.174792630 0.174792630 << 8 unaff male 65-69 UK 1998-2002
0.054956917 0.251743197 0.251743197 << 9 unaff male 70-74 UK 1998-2002
0.087542077 0.345250834 0.345250834 << 10 unaff male 75-79 UK 1998-2002
0.126170026 0.436764058 0.436764058 << 11 unaff male 80-84 UK 1998-2002
0.158273381 0.485094871 0.485094871 << 12 unaff male 85+ UK 1998-2002
0.000361579 0.013853224 0.013853224 << 13 unaff female 0-29 UK 1998-2002
0.003126994 0.070377778 0.070377778 << 14 unaff female 30-39 UK 1998-2002
0.008572049 0.155056368 0.155056368 << 15 unaff female 40-44 UK 1998-2002
0.016490198 0.256563784 0.256563784 << 16 unaff female 45-49 UK 1998-2002
0.028756473 0.374883650 0.374883650 << 17 unaff female 50-54 UK 1998-2002
0.044220151 0.488151860 0.488151860 << 18 unaff female 55-59 UK 1998-2002
0.061233051 0.575776823 0.575776823 << 19 unaff female 60-64 UK 1998-2002
0.078286465 0.636193577 0.636193577 << 20 unaff female 65-69 UK 1998-2002
0.095545853 0.681989014 0.681989014 << 21 unaff female 70-74 UK 1998-2002
0.114693025 0.718623145 0.718623145 << 22 unaff female 75-79 UK 1998-2002
0.135302716 0.739892811 0.739892811 << 23 unaff female 80-84 UK 1998-2002
0.151947021 0.744899538 0.744899538 << 24 unaff female 85+ UK 1998-2002
0.000001337 0.000106941 0.000106941 << 25 BrCa male 0-29 UK 1998-2002
0.000006959 0.000556450 0.000556450 << 26 BrCa male 30-39 UK 1998-2002
0.000010889 0.000869874 0.000869874 << 27 BrCa male 40-44 UK 1998-2002
0.000023083 0.001840075 0.001840075 << 28 BrCa male 45-49 UK 1998-2002
0.000048862 0.003872901 0.003872901 << 29 BrCa male 50-54 UK 1998-2002
0.000074754 0.005840535 0.005840535 << 30 BrCa male 55-59 UK 1998-2002
0.000110857 0.008349190 0.008349190 << 31 BrCa male 60-64 UK 1998-2002
0.000154046 0.010734333 0.010734333 << 32 BrCa male 65-69 UK 1998-2002
0.000209181 0.013686549 0.013686549 << 33 BrCa male 70-74 UK 1998-2002
0.000281452 0.016855908 0.016855908 << 34 BrCa male 75-79 UK 1998-2002
0.000317733 0.000214757 0.000214757 << 35 BrCa male 80-84 UK 1998-2002
0.000189659 0.000116019 0.000116019 << 36 BrCa male 85+ UK 1998-2002
0.000457111 0.027415880 0.027415880 << 37 BrCa female 0-29 UK 1998-2002
0.004354596 0.082488696 0.082488696 << 38 BrCa female 30-39 UK 1998-2002
0.005580613 0.078193196 0.078193196 << 39 BrCa female 40-44 UK 1998-2002
0.008737492 0.108811200 0.108811200 << 40 BrCa female 45-49 UK 1998-2002
0.013129075 0.097601856 0.097601856 << 41 BrCa female 50-54 UK 1998-2002
0.013723716 0.085408869 0.085408869 << 42 BrCa female 55-59 UK 1998-2002
0.014530921 0.043067514 0.043067514 << 43 BrCa female 60-64 UK 1998-2002
0.012251269 0.031824492 0.031824492 << 44 BrCa female 65-69 UK 1998-2002
0.013579442 0.031674024 0.031674024 << 45 BrCa female 70-74 UK 1998-2002
0.014597368 0.030789054 0.030789054 << 46 BrCa female 75-79 UK 1998-2002
0.015189253 0.004569037 0.004569037 << 47 BrCa female 80-84 UK 1998-2002
0.008885981 0.002672967 0.002672967 << 48 BrCa female 85+ UK 1998-2002
0.000000000 0.000000000 0.000000000 << 49 OvCa male 0-29 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 50 OvCa male 30-39 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 51 OvCa male 40-44 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 52 OvCa male 45-49 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 53 OvCa male 50-54 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 54 OvCa male 55-59 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 55 OvCa male 60-64 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 56 OvCa male 65-69 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 57 OvCa male 70-74 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 58 OvCa male 75-79 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 59 OvCa male 80-84 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 60 OvCa male 85+ UK 1998-2002
0.000257524 0.000257524 0.000257524 << 61 OvCa female 0-29 UK 1998-2002
0.000410633 0.002912907 0.002912907 << 62 OvCa female 30-39 UK 1998-2002
0.000429027 0.006064498 0.006064498 << 63 OvCa female 40-44 UK 1998-2002
0.000873136 0.011204105 0.011204105 << 64 OvCa female 45-49 UK 1998-2002
0.001362280 0.022947794 0.022947794 << 65 OvCa female 50-54 UK 1998-2002
0.001923589 0.027049552 0.027049552 << 66 OvCa female 55-59 UK 1998-2002
0.002457267 0.024832032 0.024832032 << 67 OvCa female 60-64 UK 1998-2002
0.002686060 0.023581526 0.023581526 << 68 OvCa female 65-69 UK 1998-2002
0.002825266 0.004555578 0.004555578 << 69 OvCa female 70-74 UK 1998-2002
0.002897527 0.004240821 0.004240821 << 70 OvCa female 75-79 UK 1998-2002
0.002942136 0.000885016 0.000885016 << 71 OvCa female 80-84 UK 1998-2002
0.001362269 0.000409780 0.000409780 << 72 OvCa female 85+ UK 1998-2002
0.000007080 0.000033773 0.000033773 << 73 PanCa male 0-29 UK 1998-2002
0.000065878 0.000314147 0.000314147 << 74 PanCa male 30-39 UK 1998-2002
0.000107852 0.000513814 0.000513814 << 75 PanCa male 40-44 UK 1998-2002
0.000250502 0.001191192 0.001191192 << 76 PanCa male 45-49 UK 1998-2002
0.000464364 0.000935538 0.000935538 << 77 PanCa male 50-54 UK 1998-2002
0.000873503 0.001736104 0.001736104 << 78 PanCa male 55-59 UK 1998-2002
0.001334669 0.002560209 0.002560209 << 79 PanCa male 60-64 UK 1998-2002
0.001981595 0.003521981 0.003521981 << 80 PanCa male 65-69 UK 1998-2002
0.002660290 0.004448589 0.004448589 << 81 PanCa male 70-74 UK 1998-2002
0.003407621 0.005230734 0.005230734 << 82 PanCa male 75-79 UK 1998-2002
0.003944167 0.002665873 0.002665873 << 83 PanCa male 80-84 UK 1998-2002
0.002279036 0.001394143 0.001394143 << 84 PanCa male 85+ UK 1998-2002
0.000008647 0.000041245 0.000041245 << 85 PanCa female 0-29 UK 1998-2002
0.000044443 0.000206249 0.000206249 << 86 PanCa female 30-39 UK 1998-2002
0.000075664 0.000321852 0.000321852 << 87 PanCa female 40-44 UK 1998-2002
0.000152430 0.000590494 0.000590494 << 88 PanCa female 45-49 UK 1998-2002
0.000310199 0.000440009 0.000440009 << 89 PanCa female 50-54 UK 1998-2002
0.000537457 0.000640837 0.000640837 << 90 PanCa female 55-59 UK 1998-2002
0.000943501 0.000923928 0.000923928 << 91 PanCa female 60-64 UK 1998-2002
0.001349378 0.001151147 0.001151147 << 92 PanCa female 65-69 UK 1998-2002
0.001961502 0.001465275 0.001465275 << 93 PanCa female 70-74 UK 1998-2002
0.002608439 0.001768415 0.001768415 << 94 PanCa female 75-79 UK 1998-2002
0.003199886 0.000962549 0.000962549 << 95 PanCa female 80-84 UK 1998-2002
0.001862729 0.000560322 0.000560322 << 96 PanCa female 85+ UK 1998-2002
0.000006142 0.000045017 0.000045017 << 97 ProCa male 0-29 UK 1998-2002
0.000010673 0.000078220 0.000078220 << 98 ProCa male 30-39 UK 1998-2002
0.000051853 0.000379627 0.000379627 << 99 ProCa male 40-44 UK 1998-2002
0.000306786 0.002240654 0.002240654 << 100 ProCa male 45-49 UK 1998-2002
0.001421401 0.010296184 0.010296184 << 101 ProCa male 50-54 UK 1998-2002
0.004466379 0.031616930 0.031616930 << 102 ProCa male 55-59 UK 1998-2002
0.010243630 0.068722198 0.068722198 << 103 ProCa male 60-64 UK 1998-2002
0.018716491 0.054352174 0.054352174 << 104 ProCa male 65-69 UK 1998-2002
0.025765479 0.069769740 0.069769740 << 105 ProCa male 70-74 UK 1998-2002
0.033055940 0.081351487 0.081351487 << 106 ProCa male 75-79 UK 1998-2002
0.036556585 0.079747356 0.079747356 << 107 ProCa male 80-84 UK 1998-2002
0.021156806 0.012942145 0.012942145 << 108 ProCa male 85+ UK 1998-2002
0.000000000 0.000000000 0.000000000 << 109 ProCa female 0-29 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 110 ProCa female 30-39 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 111 ProCa female 40-44 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 112 ProCa female 45-49 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 113 ProCa female 50-54 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 114 ProCa female 55-59 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 115 ProCa female 60-64 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 116 ProCa female 65-69 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 117 ProCa female 70-74 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 118 ProCa female 75-79 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 119 ProCa female 80-84 UK 1998-2002
0.000000000 0.000000000 0.000000000 << 120 ProCa female 85+ UK 1998-2002
If your liability classes are not in this standard format, i.e., some classes are omitted or collapsed, then you can add the --no-check option to skip the format checking, such as the example below. Below is an example Penetrance File for BRCA1 in non-standard format (notice the --no-check option). As you can see this file does not consider pancreatic cancer. This is the old version and is not used by default.
21 << --no-check --gene=BRCA1 --diseases=BrCa,OvCa --age-div=30,40,50,60,70,80 --unkn-aoo=65 --unkn-sex=1 --chr=13
0.00025 0.00314 0.00314 << 1. 0-29 unaffected female and unaffected male
0.00242 0.07307 0.07307 << 2. 30-39 unaffected female
0.01082 0.31266 0.31266 << 3. 40-49 unaffected female
0.02621 0.53921 0.53921 << 4. 50-59 unaffected female
0.04567 0.68111 0.68111 << 5. 60-69 unaffected female
0.06756 0.74146 0.74146 << 6. 70-79 unaffected female
0.09151 0.74810 0.74810 << 7. 80+ unaffected female
0.00034 0.00612 0.00612 << 8. 0-29 BrCa female and BrCa male
0.00336 0.12096 0.12096 << 9. 30-39 BrCa female
0.01130 0.35030 0.35030 << 10. 40-49 BrCa female
0.01480 0.23680 0.23680 << 11. 50-59 BrCa female
0.01800 0.19800 0.19800 << 12. 60-69 BrCa female
0.02000 0.02000 0.02000 << 13. 70-79 BrCa female
0.02390 0.02390 0.02390 << 14. 80+ BrCa female
0.00016 0.00016 0.00016 << 15. 0-29 OvCa female
0.00048 0.01824 0.01824 << 16. 30-39 OvCa female
0.00178 0.10858 0.10858 << 17. 40-49 OvCa female
0.00347 0.10410 0.10410 << 18. 50-59 OvCa female
0.00411 0.19728 0.19728 << 19. 60-69 OvCa female
0.00430 0.00430 0.00430 << 20. 70-79 OvCa female
0.00384 0.00384 0.00384 << 21. 80+ OvCa female
This website tries to read various formats whenever possible. Below are 4 formats that this website supports. If the website can read your file, a cosegregation analysis will be performed. A CPF file will be created for download. You may also want to check out my other program PedPro, which is more flexible in converting pedigree files.
Please keep in mind that if data are not collected or are misinterpreted in the first place, converting files from other formats to CPF cannot solve the problems. Also, it is very difficult to design a program to read all file formats, especially when the formats are constantly changing or highly customizable. Therefore, be prepared that this website may or may not read your format. If you need help, please email me.
1. Freestyle Format
A Pedigree File in freestyle format does not require a specific order of columns, but it should contain a header row so that the programs can identify the content of each column. Required columns are IndID Father Mother Sex Aff Age (or Liab) Proband Geno (or Allele1 and Allele2). Tabs or spaces delimit columns. Multiple delimiters are treated as one. Missing value is represented by a period (recommended) or 0 for all fields. Empty fields, space within fields, or quoted fields are not allowed. Sex, affection status, and genotypes are case-insensitive. IDs are case-sensitive. Extra columns with unrecognized headers are allowed but ignored.
PedID: Pedigree_ID. Alphanumeric. It cannot be 0. Multiple pedigrees are allowed. (See Tips 1 below).
IndID: Individual_ID. Alphanumeric. It cannot be 0. IndID does not need to be unique across pedigrees.
Father: Individual_ID of father. Put 0 for founders. If one parent is 0, both parents must be 0.
Mother: Individual_ID of mother. Put 0 for founders. If one parent is 0, both parents must be 0.
Sex: Biological sex, not the personal identification of one's own gender. 0=UnknSex, 1=M=Male, 2=F=Female.
Twin: Twin status. 0: not twin; positive integer: siblings with the same number are twins.
Aff: Affection status. Value is UnknAff (unknown), Unaff (unaffected), or a disease name (See Tips 2 below).
Age: Age-of-onset for affected and age-of-the-last-exam for unaffected individuals. 0 for unknown age.
Proband/FPTP: The first person tested positive for the variant in each pedigree. 1 for FPTP; 0 for others.
Geno: Genotype. 0=unknown, neg=negative, het=heterozygous-carrier, hom=homozygous-carrier (See Tips 3).
Allele1 Allele2: Genotype in two columns. 0=unknown, 1=reference-allele, 2=mutant-allele.
Liab: Liability class. Integers starting from 1. Zero is not allowed. This column is ignored if Age column exists.
Pop: Population. This column overrides the advanced option "Population" on the webpage.
Cohort: Year range. This column overrides the advanced option "Year range" on the webpage.
Comment: Information to be shown in pedigree drawings. (See “Tips 2” below).
Tips 1, regarding column contents: A good practice in creating an input file is to make it self-contained and self-explained. So it is better to use alphanumeric strings to name Pedigree_IDs and make it meaningful. If there is only one pedigree, the PedID column can be omitted but not recommended for the same reason. Another trick to make the file self-explained is to use English words (or at least a letter) to represent data rather than using numeric codes. For example, neg/het/hom are better than 0/1/2 for genotypes, Male/Female are better than 1/2 for sex, and the actual disease names BrCa,OvCa,PanCa are better than 2,3,4 for affection status.
Tips 2, regarding affection status: If a person is affected with multiple diseases known to be associated with the gene, then Aff should be the disease with the earliest age of onset. If a person is affected with other diseases unrelated to the associated diseases, those diseases should be ignored. You can still input the disease name in Aff and the last exam age in Age. The programs will ignore the disease and set the person as unaffected. The pedigree drawing will show this information in the comment line. If a person is affected with both an associated disease and an unrelated disease, then input the associated disease in Aff and the corresponding age of onset in Age. For cancer-associated genes, subjects are right-censored at the first diagnosis of any cancer.
Tips 3, regarding genotype: You don’t need to input genotype for obligatory carriers as the program will infer automatically. You don’t need to input negative genotype (non-carrier) for spouses even if you assume that the variant enter the pedigree only once. The program will fill in the genotype based on allele frequency. So, if the allele frequency is low, most likely the variant will enter the pedigree only once.
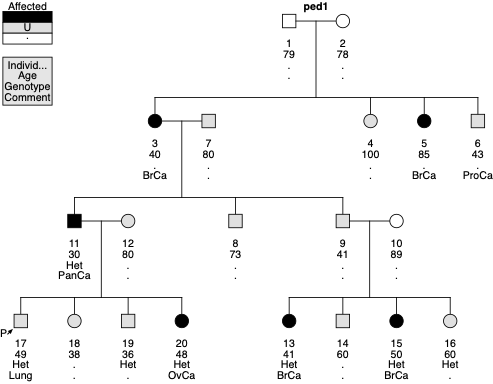
Below is an example Pedigree File and a drawing for BRCA1. Please note that affection status for analysis depends on the gene. Because the liability class model for BRCA1 involves only breast, ovarian, and pancreatic cancer, individual 6 (prostate cancer) and 17 (lung cancer) are “unaffected”. Therefore, the squares for these two persons are not filled with solid black color. Also note that the proband, individual 17, is not affected. This is correct as the definition of proband is the first person who tested positive for the variant.
PedID IndID Father Mother Sex Twin Aff Age Geno FPTP
ped1 1 0 0 M 0 . 79 . 0
ped1 2 0 0 F 0 . 78 . 0
ped1 3 1 2 F 0 BrCa 40 . 0
ped1 4 1 2 F 0 Unaff 100 . 0
ped1 5 1 2 F 0 BrCa 85 . 0
ped1 6 1 2 M 0 ProCa 43 . 0
ped1 7 0 0 M 0 Unaff 80 . 0
ped1 8 7 3 M 0 Unaff 73 . 0
ped1 9 7 3 M 0 Unaff 41 . 0
ped1 10 0 0 F 0 . 89 . 0
ped1 11 7 3 M 0 PanCa 30 Het 0
ped1 12 0 0 F 0 Unaff 80 . 0
ped1 13 9 10 F 0 BrCa 41 Het 0
ped1 14 9 10 M 0 Unaff 60 . 0
ped1 15 9 10 F 0 BrCa 50 Het 0
ped1 16 9 10 F 0 Unaff 60 Het 0
ped1 17 11 12 M 0 Lung 49 Het 1
ped1 18 11 12 F 0 Unaff 38 . 0
ped1 19 11 12 M 0 Unaff 36 Het 0
ped1 20 11 12 F 0 OvCa 48 Het 0
Below is a list of strings for the “Aff” column:
==============================================================================================================================
Name Description
------------------------------------------------------------------------------------------------------------------------------
Unaff Unaffected
Lip Lip
Tongue Tongue
Mouth Mouth
Oral Oral cavity (lip, tongue, mouth)
Saliv Salivary gland
Parotid Parotid gland
Tonsil Tonsil
Oroph Oropharynx
Nasoph Nasopharynx
Pyrifm Pyriform sinus
Hypoph Hypopharynx
Pharynx Pharynx (includes Oropharynx, Nasopharynx, Hypopharynx, Pharynx unspecified)
BCP Buccal cavity & pharynx (includes Lip, Tongue, Mouth, Saliv, Parotid, Tonsil, Pharynx)
Throat Oropharynx, Tonsil, Base of tongue
Nasal Nasal cavity and middle ear
A.sinus Accessory sinuses
Larynx Larynx
Trachea Trachea
Oesoph Oesophagus
Stomach Stomach (synonym: Gastric)
SmBowel Small intestine
Colon Colon
RS.junc Rectosigmoid junction
Rectum Rectum
CRC Colorectal cancer (includes Colon, RS.junc, Rectum)
Anus Anus
Liver Liver
Gall Gallbladder
Biliary Biliary tract
PanCa Pancreas
BilPan Biliary tract and Pancreas
Lung Lung
Thymus Thymus
Heart Heart
Bone Bone
Bone.l Bone of limbs
Bone.o Bone other than limbs
Osteo Osteosacoma
Sarcoma Soft tissue sarcoma or bone sarcoma
CM Cutaneous melanoma
NM.skin Non-melanoma of skin
Meso Mesothelioma
BrCa Breast
Vagina Vagina
Cervix Cervix uteri
Corpus Corpus uteri (Synonym: Endomet Endometrial Endometrium)
Uterus Uterus
OvCa Ovary
Penis Penis
ProCa Prostate
Testis Testis
UpUrin Upper urinary tract malignancy (kidney and renal pelvis)
Kidney Kidney
RenalCC Renal Cell Carcinoma (only for CI5-XI, a subset of UpUrin)
RCC Renal Cell Carcinoma (only for CI5-XI, a subset of UpUrin)
Ureter Ureter
Bladder Bladder
Urinary Urinary tract (includes Kidney, Renal pelvis, Ureter, Bladder, Other urinary organs)
Eye Eye
UM Uveal melanoma
Mening Meninges
CNS Central nervous system
Brain Brain
Thyroid Thyroid
MTC Medullary thyroid cancer
Adrenal Adrenal gland
Hodgkin Hodgkin lymphoma
NH.lym Non-Hodgkin lymphoma
IPD Immunoproliferative diseases
Myeloma Multiple myeloma
L.leuk Lymphoid leukemia
M.leuk Myeloid leukemia
U.leuk Cell-unspecified leukemia
Leuk Leukemia
Lymph Lymphoid neoplasms (includes Hodgkin lymphoma, Non-Hodgkin lymphoma, MALT-lymphoma, Lymphoid leukemia)
STS Soft-tissue sarcoma (Mesothelioma, Kaposi, Peripheral nerves, Peritoneum & retroperitoneum, Connective & soft tissue)
==============================================================================================================================
2. BOADICEA Format
I don’t have enough resource to catch up with the changing BOADICEA format, and I don’t have a detailed documentation about the format. Please use this format with caution.
BOADICEA v3
This web tool supports pedigree files created by the BOADICEA website. BOADICEA v3 format is automatically detected by the header line, so please do not change the header when you copy and paste from the BOADICEA website. This format is tab-delimited. The program will read column 3 (Tgt) for proband, column 1,4,5,6,7 (PedID, IndivID, FathID, MothID, Sex) for pedigree structure, column 10 (Age) for the last exam age, column 11 (YoB) for year of birth, column 12-16 (BrCa, OvCa, ProCa, PanCa) for affection status, column 18 (Mutn) for genotype, and 20-24 for breast cancer pathology (will not be used for analysis but written in the output CPF). Below is an example:
Name Tgt IndivID FathID MothID Sex Twin Status Age Yob 1BrCa 2BrCa OvCa ProCa PanCa Gtest Mutn Ashkn Er Pr Her2 Ck14 Ck56
1 Eva 1 1 2 3 F 29 1974 29 srch brca1 A +ve -ve -ve
..
BOADICEA v4 (beta)
Below is an example:
Name Tgt IndivID FathID MothID Sex MZtwin Status Age Yob 1BrCa 2BrCa OvCa ProCa PanCa Ashkn GeneticTests Pathology
1 Eva < 1 3 2 F Alive 23 1979 21 BRCA1+[gt] BRCA2–[ms] ER+ PR+ HER2–
2 mom 2 F Alive 64 1950 BRCA1–[gt] BRCA2–[ms]
3 dad 3 M Alive 65 1940
3. PROGENY Format
It should be noted that PROGENY is highly customizable. The format described here is from our institute. If you want me to adapt my programs to read your format, please send me an example file. When you click Export in PROGENY, please choose Text [tab delimited] and check the boxes “Convert newlines to spaces”, “include column headings”, and “export one row per individual”. PROGENY format is recognized if the file has a column named “Global ID” and a column named “UPN”. The file must have the following columns: Pedigree name, CreatedYr, Global ID, UPN, Mother ID, Father ID, Proband status, Gender, Genotype, Age, YoB, Cancer1, Age1. There can be multiple cancer name and cancer age columns, which should be named Cancer2, Age2, Cancer3, Age3, and so on. Here, Age# is age of diagnosis for Cancer#. The Age column is the last exam age or age of death. Program will calculate age for cosegregation analysis following these criteria: (1) if Cancer# is one of the diseases caused by the gene, age is Age#. (2) if the person is unaffected, then age is Age. If Age is missing, then calculate age from CreatedYr and YoB. Below is an example file. I formatted it to make it look good, but it should be delimited by tab, not space. Spaces are not treated as delimiters and will be converted to _ internally.
Pedigree CreatedYr Global_ID UPN Mother Father Proband Gender Genotype Age YoB Cancer1 Age1 Cancer2 Age2
TestP1 2018 2067963 2 6 7 0 M 0 . . . . . .
TestP1 2018 2067964 3 4 0 0 F 0 49 . Ovarian 27 Breast 43
TestP1 2018 2067965 4 0 0 0 F 0 . . . . . .
TestP1 2018 2067967 6 0 0 0 F 0 . . . . . .
TestP1 2018 2067968 7 0 0 0 M 0 . . Brain 50 . .
TestP1 2018 2067970 9 6 7 0 F 0 . . . . . .
TestP1 2018 2067971 10 6 7 0 F 0 . . . . . .
TestP1 2018 2067972 11 4 0 0 M 0 . . . . . .
TestP1 2018 2067973 12 4 0 0 M 0 . . . . . .
TestP1 2018 2067975 14 3 0 0 M Pos 46 . . . . .
TestP1 2018 2067976 15 3 0 0 M 0 . . . . . .
TestP1 2018 2067977 16 3 0 0 F 0 . . . . . .
TestP1 2018 2067978 17 0 0 0 F 0 . . . . . .
TestP1 2018 2067979 18 17 2 0 M 0 . . . . . .
TestP1 2018 2066941 1 3 2 1 F Pos . 1974 . . . .
4. LINKAGE Format
The website supports LINKAGE format before “makeped” (10 columns for PedID, IndivID, FathID, MothID, Sex, AffectionStatus, Liability, Proband, Allele1, Allele2) and LINKAGE format after “makeped”. Header row is allowed. Because these formats doesn’t have an age column, you need to provide a Penetrance File.
Belman S, Parsons MT, Spurdle AB, Goldgar DE, Feng BJ. Considerations in assessing germline variant pathogenicity using cosegregation analysis. Genet Med. 2020
-
1.Thompson D, Easton DF, Goldgar DE. A Full-Likelihood Method for the Evaluation of Causality of Sequence Variants from Family Data. Am J Hum Genet. 2003;73(3):652-655.
-
2.Kuchenbaecker KB et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA. 2017;317(23):2402-2416. PMID:28632866.
-
3.Antoniou AC et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer. 2008;98(8):1457-66. PMID:18349832.
-
4.Mocci E et al. Risk of pancreatic cancer in breast cancer families from the breast cancer family registry. Cancer Epidemiol Biomarkers Prev. 2013;22(5):803-11. doi: 10.1158/1055-9965.EPI-12-0195. PMID:23456555.
-
5.BOADICEA V7 Release 114.
-
6.Belman S, Parsons MT, Spurdle AB, Goldgar DE, Feng BJ. Considerations in assessing germline variant pathogenicity using cosegregation analysis. Genet Med. 2020
2020-09-18: Support BOADICEA v4 beta
2021-03-29: Allow multiple probands in one pedigree and use them all in cosegregation analysis to adjust for ascertainment. This is useful when different branches of a pedigree are ascertained separately and merged into a big pedigree.
-
1.“!!! Error: different number of columns between lines in the input pedigree file”
You pedigree file has inconsistent numbers of columns between lines. Check whether some missing values are represented by an empty string, which causes the program to skip a column. Also check whether there are some trailing empty space or tabs in some but not all lines.
-
2.“These defined events are treated as unaffected with age being the earliest event date” or
“These undefined events are treated as unaffected with age being the latest event date”
The first one is OK. That means the program recognizes the event name and knows that the person should be treated as unaffected. The latter could be a problem: the program does not recognize the event name. It is possible that you correctly input an event that is not known to the program, which is OK. But if it is a cancer site, make sure it is among one of the site names; the algorithm treats cancer and non-cancer diseases differently. Therefore, unrecognizing a cancer site may cause a different output for a cancer-related gene.
-
3.